 The Society of Biomolecular Sciences (SBS) puts on one of the most important conferences in the pharmaceutical industry. When I was working on CellKey, it was the conference where the instrument was launched and where most of the papers on CellKey were presented. So it was quite an honor to be informed that the CellKey system was awarded the 2008 Polypops Foundation Award for the Best Innovation for Microplate Design or Application by the SBS conference, and that I was one of four people selected to represent the CellKey team.
The Society of Biomolecular Sciences (SBS) puts on one of the most important conferences in the pharmaceutical industry. When I was working on CellKey, it was the conference where the instrument was launched and where most of the papers on CellKey were presented. So it was quite an honor to be informed that the CellKey system was awarded the 2008 Polypops Foundation Award for the Best Innovation for Microplate Design or Application by the SBS conference, and that I was one of four people selected to represent the CellKey team.
When I originally started at Signature BioScience in late 2000, we were investigating whether we could detect two molecules binding together by measuring the electrical impedance of the system and calculating permittivity. Detecting binding is important for drug discovery because most drug candidates (aka agonists) work by binding to a receptor molecule on the surface of a cell, like a key going into a lock. Detecting binding events in other instrument systems involves finding a fluorescent tag, which you attach to the agonist, and then hope that the fluorescent tag doesn’t disrupt the way in which the agonist binds to the receptor.
Signature BioScience was funded on the promise of being able to detect these binding events without having to use any sort of extraneous tag, instead detecting binding by measuring the electrical impedance of a system that only included agonist and receptor molecules. The original idea was to be able to detect binding events of molecules in solution, with no messy cells or biological material interfering. Unfortunately, a year and a half later, we discovered that the changes were just too small – we were trying to measure events out in the sixth decimal place of permittivity, which was impossible. Permittivity is temperature dependent, and the effects we were measuring were so small that even though we eventually got our detector to be temperature stable to within a ten thousandth of a degree (0.0001 degrees), we actually needed to be stable to a millionth of a degree.
However, while most of the company was going down the route of measuring molecular binding, Vivian Liu (a cell biologist) and Chris Fuller (an electrical engineer) decided to experiment with using our technology to measure what the bioimpedance would look like if we added an agonist to an actual cell culture. In other words, rather than isolating a single receptor molecule and measuring the binding directly, they cultured cells to “overexpress” surface receptors of a single type, and then added different drugs to see what happened. I got called in because they needed software to run the experiment and to analyze the results.
The results were promising. We couldn’t quite figure out what the data meant, but we could definitely see a difference in the data that we got when we added a drug that bound to the receptor versus one that didn’t, and we could even see different characteristics depending on the type of cell pathway being activated. It was enough that we got approval to do a more systematic proof of concept study (well, actually, we were doing all of this under the radar, at least on the engineering side – we were supposed to be working on the big molecular project). We set up a few different cell lines, and “screened” them against a panel of agonist molecules to see what happened. We didn’t have a lot of statistics (at the time, we were using plates that had only two wells, so we had a single control well and a single experimental well), but things seemed to be shaping up.
Signature eventually went bankrupt in April of 2003 due to a variety of factors, including the CEO making some poor decisions (which he later reprised at his next company). Before I started looking for another job, MDS Sciex, a company that had been partnered with Signature to commercialize the technology we were developing, got in touch. They thought that the cell data from our proof of concept study was promising enough that they wanted to continue working on it. So despite MDS Sciex being a mass spectrometer company based in Toronto, they hired 12 of us Signature refugees in San Francisco to continue working on this technology.
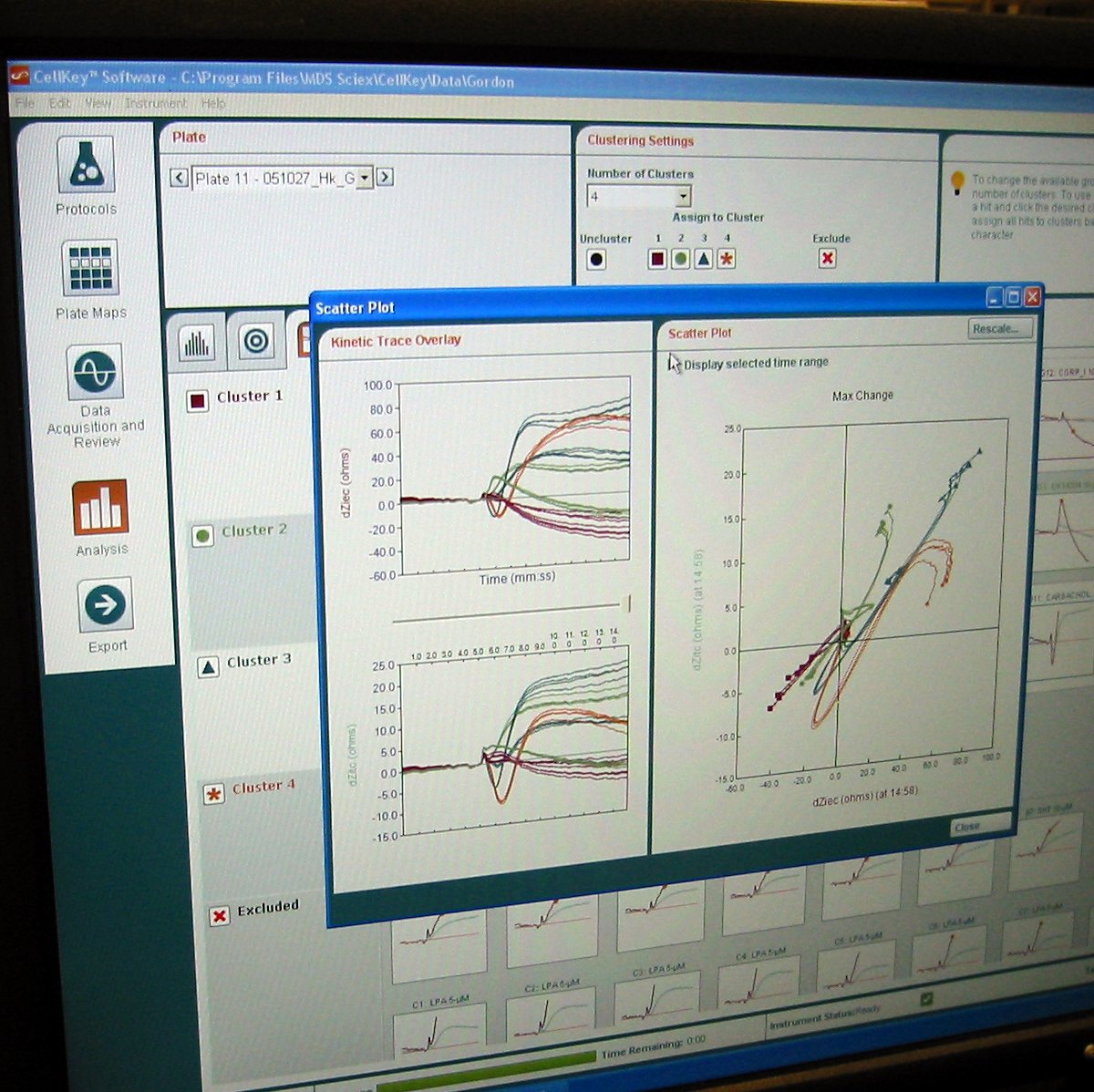 Over the next two years, we studied the phenomenon far more closely. We learned that what we were seeing in our results was the ways in which the cell changed in response to the initial binding event. The cell was essentially acting as a biological amplifier for the binding event, creating effects big enough for us to see and measure consistently. We figured out how to mathematically characterize the different signals we were seeing so that we could give biologists meaningful results; instead of 60-dimensional bioimpedance data, the program could spit out “This looks like a Gq pathway”. We worked with partner companies to test out the technology with their experiments and passed all of the tests successfully. The technology was scaled up and released as the CellKey instrument in September 2005, and it was incredibly satisfying to see the finished product.
Over the next two years, we studied the phenomenon far more closely. We learned that what we were seeing in our results was the ways in which the cell changed in response to the initial binding event. The cell was essentially acting as a biological amplifier for the binding event, creating effects big enough for us to see and measure consistently. We figured out how to mathematically characterize the different signals we were seeing so that we could give biologists meaningful results; instead of 60-dimensional bioimpedance data, the program could spit out “This looks like a Gq pathway”. We worked with partner companies to test out the technology with their experiments and passed all of the tests successfully. The technology was scaled up and released as the CellKey instrument in September 2005, and it was incredibly satisfying to see the finished product.
Getting the SBS Award brought all of those memories back, so I thought I’d share them. I don’t think I’ve ever chronicled the four years I spent working on this project, and figured this would be a good excuse. It was a really fun project to work on, melding biology, physics, engineering and software. I also learned a lot from the experience, from team building to language alignment to identifying the real problem. And it feels good for the four years I spent on that project to be validated in some small way by the SBS Award.

Hello eric
I am actually in the sciences and stumbled across your blog. Its amazing all of the things that you have done. I was interested in the cellkey technology and wanted to know if there was anyone in the Midwest that had one of these, or just on the coasts?